
Also, the hierarchical information (domain divided into elements, each element contains several points) is lost. Also, the solution can be defined twice on the cell surfaces, as the DG framework allows discontinuous solutions.

Pro: All plugins that work with the standard VTK unstructured cell types will continue to work without changing anything.Ĭon: Since the integration points are not distributed evenly, it might be difficult to find the correct location of the vertices. Use one voxel for each integration point. Several possible ways come to my mind, but I do not know which one is the most promising: My question is whether anyone has experience with visualizing such data efficiently with ParaView/VTK, and what approach you chose to represent the data in VTK. As opposed to finite volume methods, within each cell there is not just one value for the solution vector $\mathbf$ at multiple Gauss integration points. Similarly to finite volume methods, the problem domain is divided into cube-shaped cells ("elements"). Many of the ParaView examples use Show() Render(), which generates a frozen, non-interactive window.I would like to visualize simulation results, obtained using the discontinuous Galerkin (DG) approach, within ParaView.
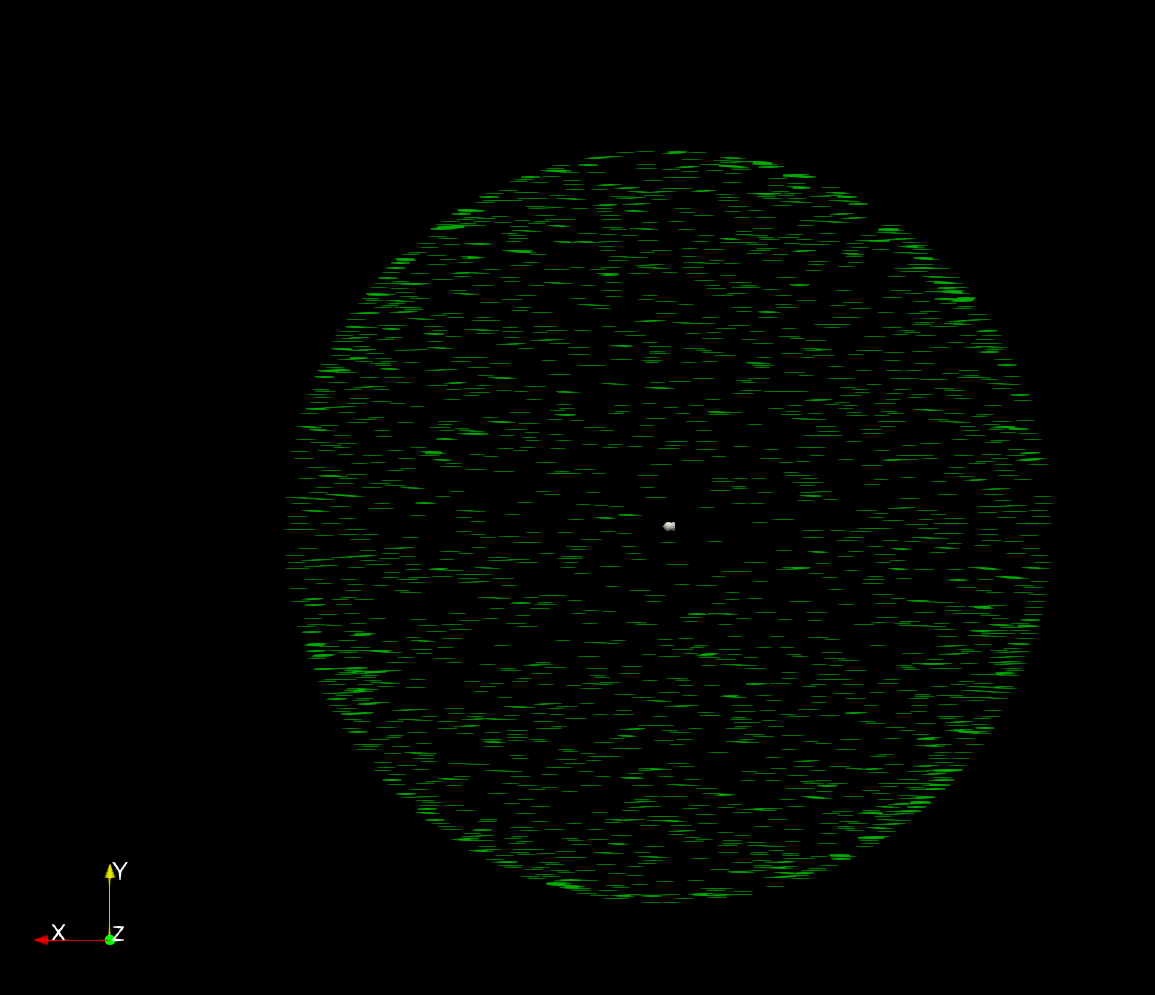
If your computer has a discrete GPU, 1024 or more points per dimension may be possible, with proportionately slower rendering.Ī key feature of ParaView is that it gracefully degrades to render 3D output even on modest devices, as long as you don't select too large Sampling Dimensions. If your computer does not have a discrete GPU, using more than about 256 grid points in a dimension may crash ParaView, even with 16 GB RAM. The data can be sampled with Property "Sampling Dimensions" at higher resolution, but this takes proportionately more GPU resources. Then, in the Properties browser, select Representation: Volume. nc filename, Add Filter, Resample as Image In the "Pipeline Browser" right click the. The data needs to be filtered to display as a volume in ParaView. The "convert_data" scripts store the grid in the NetCDF file. To use a grid, which is particularly important for non-uniform gridded ionospheric data, store the NetCDF4 data with the grid data embedded in the file. ParaView can load single-variable NetCDF4 files. In the ParaView GUI, select "Volume" and the variable name to actually render the data. However, non-COARDS, non-CF single variable files can be loaded with the plain NetCDF filter.
Paraview vtk write matlab hangs windows#
Windows Gemini users are already using MS-MPI, so they should also use the Windows ParaView MPI installer. MacOS and Linux have only MPI ParaView downloads. The example using cuml "UMAP MNIST Example.ipynb" requires a Linux system with GPU due toĬuml is a general machine learning GPU library unrelated to IPyParaView.Īlso the Dask example "Dask-MPI_Volume_Render" is for running on Dask-MPI HPC cluster, which would need to be setup by your IT staff perhaps. Iso-Surfaces_with_RTX.ipynb "RTX" requires an appropriate GPUĪdvanced examples (not for beginning user).

We suggest examples below, which do not require a discrete GPU (they run on a light laptop): And use the web browser Jupyter Notebook that automatically opens to browse and run the examples.


 0 kommentar(er)
0 kommentar(er)
